CAUSALPATH – ABOUT
Discovering the causal mechanisms of a complex system of interacting components is necessary in order to control it. Computational Causal Discovery (CD) is a field that offers the potential to discover causal relations under certain conditions from observational data alone or with a limited number of interventions/manipulations.
An important, challenging biological problem that may take decades of experimental work is the induction of biological cellular pathways; pathways are informal causal models indispensable in biological research and drug design. Recent exciting advances in flow/mass cytometry biotechnology allow the generation of large-sample datasets containing measurements on single cells, thus setting the problem of pathway learning suitable for CD methods.
CAUSALPATH builds upon and further advances recent breakthrough developments in CD methods to enable the induction of biological pathways from cytometry and other omics data. As a testbed problem we focus on the differentiation of human T-cells; these are involved in autoimmune and inflammatory diseases, as well as cancer and thus, are targets of new drug development for a range of chronic dis-eases. The biological problem acts as our campus for general novel formalisms, practical algorithms, and useful tools development, pointing to fundamental CD problems: presence of feedback cycles, presence of latent confounding variables, CD from time-course data, Integrative Causal Analysis (INCA) of heterogeneous datasets and others.
Features that complement CAUSALPATH’s approach:
- methods development which will co-evolve with biological wet-lab experiments periodically testing the algorithmic postulates,
- Open-source tools will be developed for the non-expert, and
CAUSALPATH brings together an interdisciplinary team, committed to this vision. It builds upon our group’s recent important results on INCA algorithms.
The Role of Causal Discovery: Causal knowledge is the ultimate goal of any scientific endeavour. Causal modelling goes beyond traditional statistical modelling by allowing one to predict the effects of actions and interventions on a system, e.g., the effects of changing a web interface, prescribing a new medication, or designing a new drug. Traditional means of inducing causal relations demand a manipulation (perturbation, intervention) to be performed on the system. In contrast, Computational Causal Discovery (CD) methods argue that given certain broad assumptions about the nature of causality one can induce causal relations from observational data only or a limited number of manipulations / interventions. One can then analyse archived data, forgoing expensive, time-consuming, or even impossible experiments, and determine certain of the causal mechanisms. In the era of Big Data, with increased volume and variety, such ideas have the potential to revolutionize data-analysis and science in general.
The Role of Biological Pathways: Biological Pathways are defined as “a series of actions among molecules in a cell that leads to a certain product or a change in a cell (National Human Genome Research Institute). An example of a pathway involved in signaling transduction in T-cells is shown in Figure 1. To a computer scientist, pathways are informal causal models, involving genes, proteins, metabolites, and other molecules. Pathways are often qualitative (except some well-characterized metabolic pathways): the functional form of the interactions is often not captured. Due to complexity of cells there is a high degree of uncertainty of the validity and completeness of the current pathways, and the methods of discovering novel pathway is far from satisfactory. Yet, they are indispensable and ubiquitous in molecular biological research and drug design as different activation of the same pathway may lead to different cell-fates. A large community and industry has grown around pathways: dozens of other public, as well as commercial pathway knowledge bases, standards for communicate them, software tools for constructing them, visualizing them, and juxtaposing with data, and statistical methods for employing them during omics data analysis (Varadan et al. 2012; Viswanathan et al. 2008; P D Karp et al. 1999). Arguably, biological pathways consist of the distilled knowledge of a large part of molecular biolo-gy regarding the inner mechanisms of the cells.
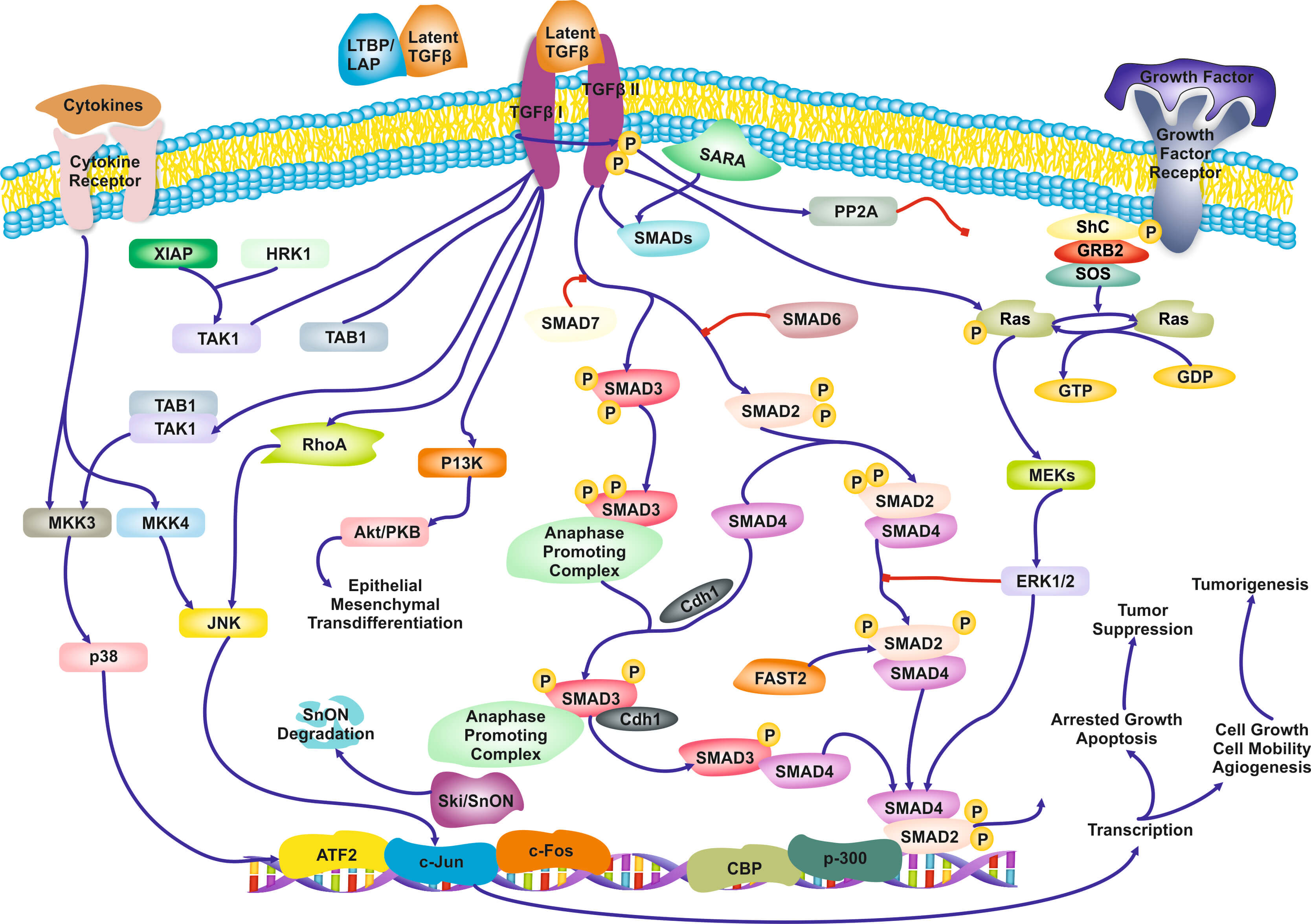
Figure 1. Qualitative, partial pathway involved in TGF-β signaling.
Learning Biological Pathways: The literature on pathway learning is significant (Van Riel 2006; Opgen-Rhein & Strimmer 2007; Markowetz & Spang 2007; Werhli et al. 2006; Yoshida et al. 2008; Varadan et al. 2012; O. D. Perez & Nolan 2006; Viswanathan et al. 2008; K. Zhang et al. 2012; Chou & Voit 2009; D Pe’er et al. 2001; Ong et al. 2002; Dale et al. 2010). In summary, there are two approaches for learning biological causal networks. The first one employs dynamic systems modeling such as Ordinary Differential Equations to derive a fine-grained, detailed mechanistic model (Yoshida et al. 2008). While quite powerful, such methods assume the structure and the equations governing the pathway are known in detail and essentially focus on estimating the parameters. Perhaps the two major problems with this approach are the inherent inability to handle latent variables and difficulty of scaling up. We consider this a synergistic approach that can be employed after a portion of the structure of a pathway has been identified. The second approach is based on graphical and causal probabilistic models, particularly the simplest and most causally-naïve models: Bayesian Networks (D Pe’er et al. 2001) and Dynamic Bayesian Networks (Ong et al. 2002). Intricate issues such as the effects of data discretization, the effects of violations of Gaussianity of errors or linearity of relations or other statistical assumptions made, the robustness to finite sample size, model equivalences, the presence of feedback cycles, possible confounding effects, low-sample estimation effects, multiple testing effects, and others are typically ignored. Some of these problems led one of the pioneering, founding teams of the field to declare the feat of network induction is improbable with aggregate (non single-cell) data (Chu et al. 2003). Considering multiple datasets, prior knowledge, and temporal data simultaneously is beyond current methods. To our knowledge, none of the more sophisticated CD methods has been successfully applied on this problem. As a rule, existing methods are proof-of-concept exercises, not validated experimentally (Dale et al. 2010; Ong et al. 2002) and haven’t penetrated biological research methodology. The pathways biologists rely upon are still manually constructed and curated (Varadan et al. 2012) provides a review for some of these methods). Therefore, learning pathways is considered an important unsolved problem that requires fundamental, formal, principled, rigorous and significant extensions of CD to solve it.
Cytometry Technologies: Cytometry is a rapidly evolving, highly sophisticated biotechnology. The basic ideas behind it however, are conceptually simple: in cytometry a selected set of molecular quantities (e.g., a set of proteins of interest) is somehow labeled / tagged in every cell of a biological sample (e.g., blood). In flow cytometry the labels are fluorescent chemicals that can re-emit light upon light excitation; their concentration can be measured with lasers. In mass cytometry the labels are rare isotopes, normally not found in living cells; they are shot into plasma, ionized and a spectrometer measures their concentration. The main advantages of cytometry are that it can measure the concentrations of the tagged quantities (a) in single cells at a rate of (b) a few thousand cells per second. Thus, sample sizes in the thousands are common. This provides unprecedented opportunities for CD methods, as well as other sample-hungry statistical and mathematical modeling techniques. In contrast, all other omics technologies (e.g., microarray gene expressions, Next Generation Sequencing data, mass spectroscopy, methylation arrays, etc.) measure the average of molecular quantities in cultures of millions of cells, often at a different stage or even of different cell type. The averaging effect of these other technologies is a phenomenon that severely affects the ability for CD methods to correctly model a system (Chu et al. 2003). Cell signaling and communication information is lost. Pathway dysregulation information due to disease is lost. Cell/tissue type differentiation information is lost.
Integrative Causal Analysis (INCA) to Overcome Technical Limitations: A serious disadvantage of cytometry technologies is that for the foreseeable future they can only measure up to 100 variables simultaneously (Bodenmiller et al. 2012) hindering the discovery of the role of unsuspected, unmeasured factors (most flow cytometry current-ly measure fewer than 12 variables at a time). A recent new CD paradigm named INCA (Tsamardinos et al. 2012) may provide a way to overcome the limitations. INCA methods can learn from datasets measuring overlapping variables. Thus, causal models can be constructed from multiple runs of a cytometers, each time measuring different quantities. INCA methods also deal with datasets under different experimental conditions, which is also important to refine the causal direction of relations.
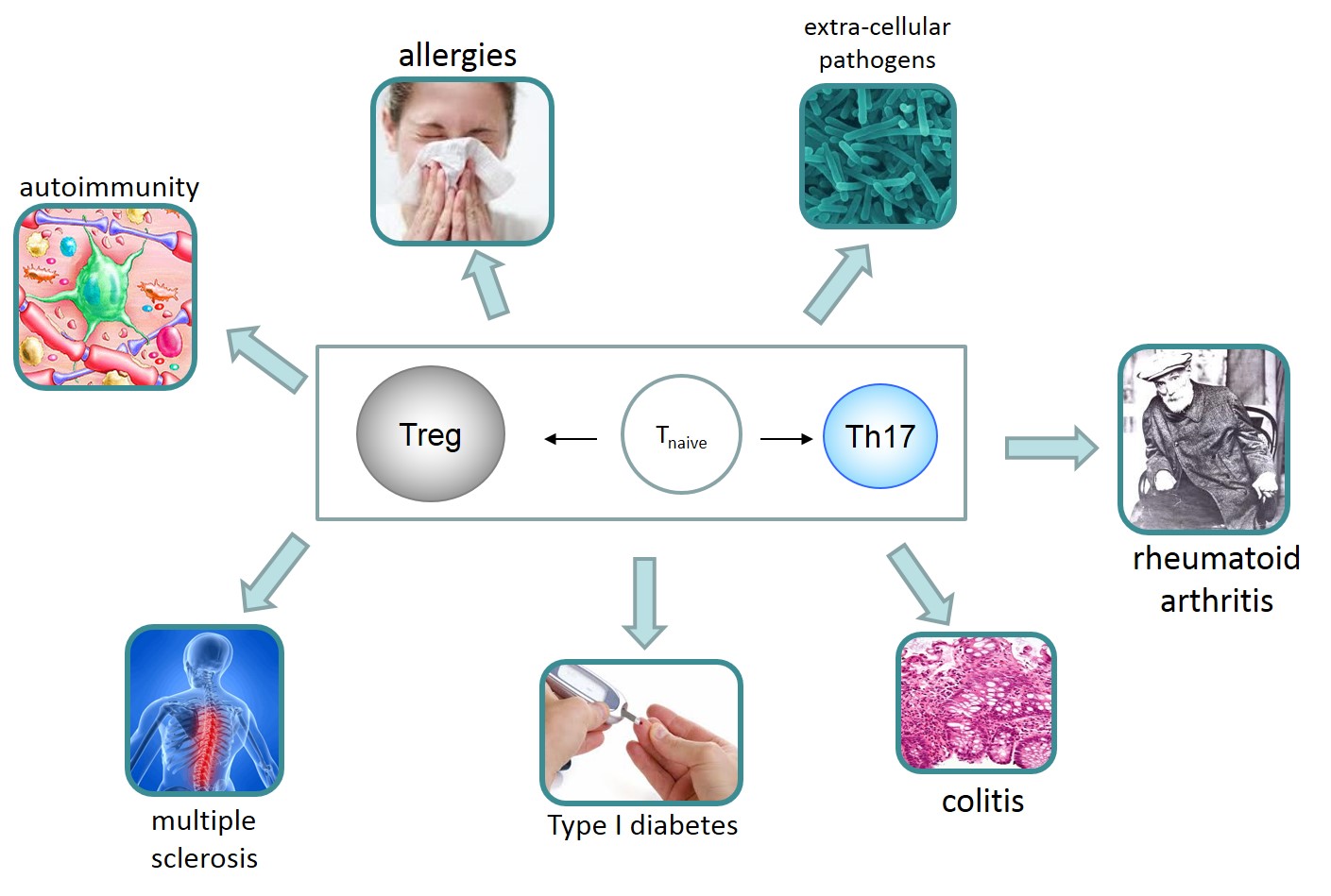
Biological Test Bed: The specific system under study is the human T helper (Th) cell and the causal mechanisms governing the differentiation and the interaction between T regulator (Treg) and conventional T human cells (specifically, T helper 17 cells, Th17). Dysregulation / differentiation imbalance of these cells is critically involved in several autoimmune and inflammatory diseases, allergies, and infection induced pathology (Buckner 2011) (e.g., multiple sclerosis, colitis, rheumatoid arthritis, Type 1 diabetes). As such, they are interesting for drug development.
Inducing (quantitative) pathways from data is arguably the Holy Grail of molecular biology. It corresponds to reverse engineering of the mechanisms of the cell, allowing to discover useful genetic modifications to organisms, designing drugs to treat diseases, planning of optimal therapies, or predicting the development of disease. Construction of pathways is typically a manual, labour-intensive exercise that is typically based on retrieving results from the literature corresponding to decades of experimental work. In addition, it is prone to errors of commission, but mostly on omission of unsuspected interactions, interactions among different pathways, interactions with environmental factors, or dysregulations due to disease. The existing knowledge bases contain pathways that are often generic, in the sense that they do not capture interactions that are context or patient dependent (Varadan et al. 2012). In addition, the effect of new drugs on the pathways is often unknown or only qualitatively known. Thus, pathway induction on a patient-specific base is necessary for true personalized medicine and understanding of complex diseases such as cancer: “single-cell analyses provide system-wide views of immune signaling against which drug action and disease can be compared for mechanistic studies and pharmacologic intervention” (Bendall et al. 2011). This challenge provides CAUSALPATH with a dimension that is rare in CD research.
To such an end, cytometry poses as a promising tool. In fact, several breakthroughs both in cytometry (and other biotechnologies) and CD methods are fast converging to revolutionize the field. The list of measurable quantities in cytometry technologies grows every day; morphological characteristics of the cell, hundreds of different protein expressions as well as proteins’ modifications can now be measured. Multiplexing is a technique that facilitates measurements under dozens of different perturbations simultaneously (Bodenmiller et al. 2012). Technologies such as high-content image-based cytometry (De Vos et al. 2010) are steadily increasing their throughput and can currently measure the sub-cellular localization of the variables, thus soon adding a spatial dimension to the data. A wealth of cytometry data is becoming publicly available for analysis at a growing pace: just the dataset from (Bodenmiller et al. 2012) contain more than 100 million (!) samples/cells from 8 different human donors, under the influence of different drugs in different dosages at different times and several experimental conditions.
In parallel and equally important, CD methods are getting ready to tackle the new challenges. The mainstream CD approach relies on analysis of conditional dependencies and independencies in the data distribution pioneered by groups centered on J. Pearl (Pearl 2009) and the Glymour, Scheines, Spirtes groups (Spirtes et al. 2001). These advances were partly responsible for Prof. Pearl’s Turing Award in 2011. But, novel directions are also being explored. First, fundamentally new principles for identifying causal relations have been discovered. In essence, these principles identify and exploit data distribution characteristics that are asymmetric w.r.t. the causal direction. For example, the structures X→Y and X←Y are distinguishable from an observed distribution if the relation is linear but the noise terms are non-Gaussian (S Shimizu et al. 2006; A. Hyvärinen et al. 2008) or the relation is non-linear and the noise is Gaussian (P. Hoyer et al. 2009). Second, formalisms (such as semi-Markovian models and Maximal Ancestral Graphs) and respective algorithms that admit the presence of latent confounding variables are advancing to maturity (Leray et al. 2008). The ability to handle the effect of latent variables is paramount in most real settings and biology in particular and was the source of major criticism of early work in CD. Third, methods for learning causal models from multiple heterogeneous datasets or knowledge sources have also appeared. Our group has coined the term INCA to collectively characterize them (Tsamardinos et al. 2012). Such methods include learning causal models from datasets obtained under different experimental conditions (perturbations) (Hauser & Bühlmann 2011). As mentioned above, INCA algorithms may be necessary to overcome the technical limitations of cytometers. In addition, they may allow cytometry data to be coanalyzed with other omics public datasets. Figure 2 graphically depicts an illustrative case-scenario.
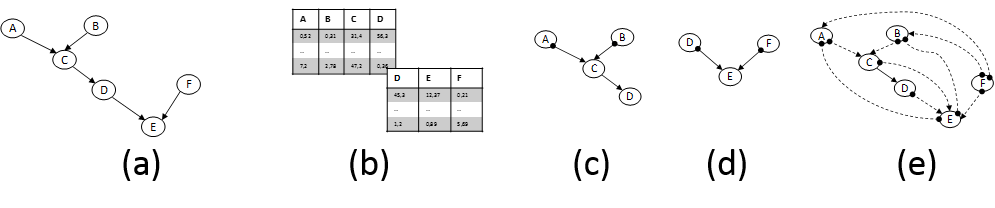
Figure 2. (a) Fictional pathway. Edges represent direct causal relations; the parameters determining effect sizes are not shown. (b) Flow-cytometry datasets measuring overlapping variable sets. (c) and (d) Partially-oriented Acyclic Graph models induced by analysing each dataset in isolation under typical assumptions. Edges denote: → causation and •→ causation plus possible confounding. (e) Pairwise Causal Graph induced by co-analysing both datasets with recent methods developed by the PI’s group that can deal with datasets over overlapping variables. The solid edge C→D means all models that fit both datasets agree on its presence; dashed edges imply there are equivalent models with and without the edge. Notice that non-trivial inferences are drawn: C and F are found not causing each other even though they are never measured together. The model can be employed to bound the effects of intervening to some of the quantities, thus suggesting possible drug targets.
Objective 1:
Robust, Conservative Causal Discovery, Characterization of Confidence and Structural Uncertainty of Models: develop algorithms that are robust to violations of assumptions and rank causal claims according to their confidence.
Objective 2:
Query-driven, Local, and Opportunistic Causal Discovery: develop algorithms that focus on a causal feature of interest (e.g. how to best affect / control variable X) or a local / partial model instead of complete models; focus on learning the parts of the system for which we can be most confident about.
Objective 3:
Causal Discovery with Feedback cycles, Time-Course Data, and Latent Variables: invent novel formalisms and algorithms to simultaneously deal with all the above characteristics.
Objective 4:
Incorporation of Rich Prior Knowledge into Causal Discovery: invent formal languages and respective algorithms that can represent and reason with the wealth of existing biological knowledge, such as known transcription factors, generic pathways models, and others.
Objective 5:
Integrative Causal Analysis from Multiple Experiments and Overlapping Variable Sets: extend existing algorithms to robust and practical settings for application to cytometry data coupled with other omics datasets (e.g., gene expression).
Objective 6:
Planning for Optimal Next Experiments: invent a formal language to represent the types of experiments / manipulations that are possible, their cost, as well as the goals of the user and corresponding algorithms that, based on the currently known causal knowledge, find the optimal plan of actions.
Objective 7:
Induction of Th17/Treg differentiation and interaction pathways: Objectives 1-6 will first be applied, refined, and tested on public datasets (Bodenmiller et al. 2012). Subsequently, they will be applied on newly produced, unpublished data related to two specific biological problems with direct impact to human health:
- elucidating the role of the TGF-β factor in Th17/Treg differentiation pathway dysregulation and
- unravelling the biological mechanisms that are at the basis of Treg cells’ immunosuppressive activity towards conventional T helper cell signaling.
Objective 8:
Open-Software, Open-Architecture Causal Discovery Tools: design an open software architecture that can allow plug-and-play capabilities to new algorithms (allowing crowd-sourcing them) as well as causal discovery tools for the non-expert.
Objective 9:
Dissemination and Exploitation: disseminate through publications, newsletter, tutorials, conference and workshop organizations, project web-pages, demos, software tools; exploitation of research results in the form of software licenses and systems may also be sought.
- Computational research centered around a specific biological problem: CD methods in Computer Science measure four decades of de-velopment, heated debating and controversy, generation of widely applicable new paradigms such as reasoning with Bayesian Networks, as well as a recent wealth of successes (some are mentioned below). Yet, when trying to advance the field in novel directions, one runs the risk of making mathematically convenient, but unrealistic or impractical assumptions about the causal nature of the system under study. In the proposed work, a different approach is taken: a specific, well-defined biological problem is set forth to focus methodological research to-wards directions that lead to practical results. Yet, it is expected that solving such a challenging problem will also result in fundamental ad-vances in CD methods that transfer to many other domains and impact CD throughout: advances such as the ability to handle feedback cy-cles, to admit latent confounding variables, to characterize the confi-dence of discoveries, to perform CD from time-course data, to do Inte-grative Causal Analysis (INCA) of heterogeneous datasets, plan future experiments and others. Pathway induction has been selected both for its potential impact to biology, drug discovery, and personalized medi-cine as well as the opportunities for advancing CD methods.
- Interdisciplinary, experimental research: the algorithmic and theory de-velopment will co-evolve with biological wet-lab experiments periodi-cally testing the algorithmic postulates and predictions; the PI’s prestig-ious collaborators in Karolinska Institutet, fully equipped with a flow-cytometry facility (currently applying for funding for a mass cytometer too), will provide the biological expertise, performance of biological experiments, and interpretation of results.
- Open-source tools for CD will be developed for the non-expert, with an open architecture for crowd-sourcing solutions.
- Exploitation of the results could potentially be sought out, particularly to (a) cytometry manufacturers, (b) bioinformatics software companies, as well as (c) general data-analysis software companies in need to CD methods.
Apart from the benefit in understanding Th17-associated diseases CAUSALPATH primarily aims for a major impact to computer science. First, the test-bed setting of the biological problem will focus a concerted effort on the part CD community. A collection of related datasets, causal discovery tools, and new biological data including experiments testing predictions will instigate further research. Second, as computer scientists we strive for general algorithms, applicable to a number of domains. The algorithms proposed for development involve fundamental problems in CD. As evidence of generality, we point to our work on INCA methods in (Tsamardinos et al. 2012) applied and robustly performing in 20 different domains from financing and drug design to text analysis. Third, successful completion of the project will further establish CD as realistic, practical, and useful in the eyes of the data-analysis community. Current perception ranges from the “CD over-enthusiasts” (learn a Bayesian Network and interpret each edge X→Y in the network causally, ignoring issues of model equivalence, estimation fluctuations, possible confounding and assumptions’ violations), to the “CD sceptics” (“association is not causation”, period).
If successful, CAUSALPATH will impact science in several ways and create new opportunities for science. The algorithms proposed for development involve funda-mental problems in CD. The Objectives set forth will
- bring awareness of the potential and comparative benefits of CD,
- increase confidence in the predictions of the methods,
- increase understanding of the methods and comprehension of the results,
- increase the availability of end-to-end analysis CD protocols and related tools.
New directions in CD methods will open, particularly with respect to integrative analysis. The need for semantic annotation of datasets with meta-data about their collection, what they measure, how they measure it, and other characteristics that allow the automatic semantic retrieval and large-scale, joint analysis will become evident. The availability of robust, practical CD methods, as well as easy-to-use tools for non-experts could support discoveries in all of natural sciences. Of course, the largest, immediate impact is intended for biology where it could boost our understanding of cellular mechanisms and their dysregulations related to disease.
CAUSALPATH aspires to contribute a first, solid step towards materializing the following futuristic scenario:
“It is 2030 and the highly anticipated Pathway Cytometer X machine is ready for mass production after extensive testing (Objective 7). A slot allows the deposit of biological samples of tissues; a user interface allows the researcher to dictate a set of known molecular quantities (e.g., proteins) among which they are interested in discovering their interconnecting causal mechanisms. Α range of conditions and a time horizon can also be input and further direct the experiment. Subsequently, the machine subjects the samples to a series of manipulations (e.g., in-duction of biological signals) and measures the concentrations of only a 100 (the maximum possible) out of all possible molecular quantities in tens of thousands of single cells and at different time-points. Sophisticated Causal Discovery (CD) software (Objectives 8-9) internally creates a first rough draft of the causal mechanisms taking place (Objectives 1-3). The machine downloads all related prior knowledge from public knowledge bases, as well as related omics datasets in public repositories. The incorporation of this knowledge and co-analysis with the public data help the CD software further refine the causal model (Objectives 4-5). The CD software determines the next-best-experiment for the machine (Objective 6): this may include measuring next a different (but overlapping) set of 100 variables so the model is expanded, measuring under different manipulations so the causal direction in uncertain cases is refined, or measuring at different time-points so the dynamics are better determined. The CD software co-analyses the new data with all previous ones (Objectives 4-5). The machine repeats the cycle for as long as the time and cost constraints are respected. In the end, a formal representation of causal relations, their strength, parameters, and functional form, as well as their structural uncertainty (e.g., presence or direction) and parametric uncertainty is output.
The machine is a big success! Diagnostic centres around the world use it regularly to detect specific deregulations in tumour cells and select the optimal treatment, on a personal level. Pharmaceutical companies are using the models to test in silico new drugs or design new ones. It helps biologists around the world better understand cell mechanisms in a variety of tissues and species.
At the same time, in the IT-world CD software has become an integral part of a data-analysts arsenal. Confidence and acceptance in the methods has increased (Objectives 8-9); after all, it does lead to new biological discoveries every day. Automated or semi-automated advanced CD tools are employed in all natural sciences. Causal analysis is now the main means by which models learnt from one or more datasets transfers to other contexts and situations (when possible) (Pearl & Bareinboim 2012). But most importantly, Integrative Causal Analysis (INCA)(1) methods have allowed persistent and continuous learning. Large centres exist with the task to continuously learn and update their models and knowledge by incorporating and co-analysing every new study with previous ones(2). The learnt models are formal: the system can explain the conclusions and beliefs of the models and help a human expert retrace, verify, and visualize them leading to evidence-based science.”
(1) J. Pearl has independently coined the name Meta-Synthesis (Pearl 2012) for the same purpose as INCA a few months after our publication (Ioannis Tsamardinos et al. 2012). The term Meta-Synthesis has also other meanings in the statistical literature (Stern & Harris 1985) and thus its overloading may be confusing.
(2) A US-based effort for continuous learning led by Carnegie Mellon University and funded by DARPA, Google, and NSF has already begun named Never-Ending Language Learning (NELL). NELL however, is quite different in methods as well as goals: it learns semantic rules from documents and is not – at the moment – applicable to scientific measurement data.